NBLtools
Diffusion MRI Preprocessing Pipeline

Diffusion MRI Preprocessing Pipeline

NBLtools is a comprehensive preprocessing pipeline for diffusion MRI data. It provides automated tools for data quality control and correction, ensuring your diffusion data is optimally prepared for analysis.
There are two ways to install and use NBLtools:
Detailed installation instructions for both methods are available below on this page.
Before running NBLtools, organize your data with the following structure:
subject01/
└── dwi/
├── dwi.nii.gz # Diffusion weighted images
├── dwi.bval # B-values file
├── dwi.bvec # B-vectors file
├── dwi_rev.nii.gz # Reverse phase encode data (optional)
├── dwi_rev.bval # B-values for reverse phase
├── dwi_rev.bvec # B-vectors for reverse phase
└── acqparams.txt # Acquisition parameters
The acqparams.txt file should contain the phase encoding directions and total readout time:
0 1 0 0.086 0 -1 0 0.086
Each row corresponds to a volume: phase encoding direction (x y z) and total readout time in seconds.
Once your data is organized, process it with a single command:
run_all_tu ./subject01 -f -p -b 1 -i -t -m -e -c 0 -q
Each parameter controls a specific preprocessing step:
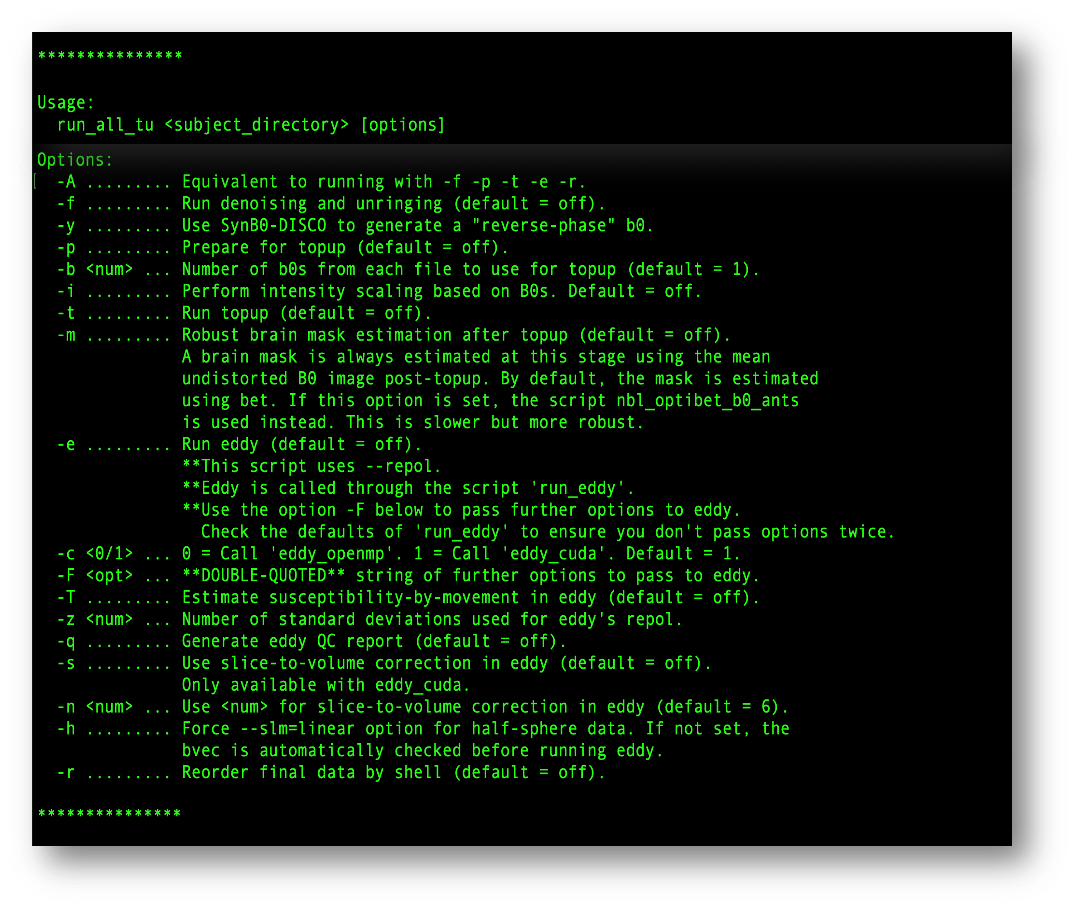
The screenshot above shows all available parameters and their descriptions directly from the command line help.
For users who want to install NBLtools directly on their Linux, Windows Subsystem for Linux (WSL), or macOS system.
NBLtools requires the following software dependencies to be installed:
Advanced Normalization Tools for image registration and segmentation
FMRIB Software Library for brain imaging analysis
Tolerably Obsessive Registration and Tensor Optimization Indolent Software Ensemble
Minimum: 8GB
Recommended: 16GB+ for large datasets
After installing the dependencies, you need to add them to your PATH. Edit your ~/.bashrc file (Linux/WSL) or ~/.bash_profile (macOS) and add these lines:
# Add NBLtools to PATH (including all subdirectories) NBLTOOLS_DIR=/path/to/your/nbltools # Change to your installation path if [ -d "$NBLTOOLS_DIR" ]; then NBLTOOLS_PATHS=$(find $NBLTOOLS_DIR -type d ! -path "*.git/*" | tr '\n' ':') export PATH="$PATH:$NBLTOOLS_PATHS" fi # Example for ANTs (simple path addition) export PATH="/path/to/ANTs/bin:$PATH" # For FSL and TORTOISE, follow their specific installation instructions
Note:
• Change NBLTOOLS_DIR to where you cloned/installed NBLtools
• For ANTs, replace /path/to/ANTs/bin with your actual ANTs installation path
• FSL and TORTOISE have specific setup scripts - follow their official installation guides
After saving the file, reload it by running: source ~/.bashrc
For Windows, Linux, and macOS (Intel) users, we provide a Docker container with all dependencies pre-installed. This is the easiest way to get started with NBLtools.
Run NBLtools interactively with all dependencies included:
Replace /path/to/your/data with the actual path to your diffusion MRI data directory. Your data will be accessible inside the container at /data
For questions, bug reports, or feature requests: